PyMOL
www
PyMOL is computer software, a molecular visualization system created by Warren Lyford DeLano. It is user-sponsored, open-source software, released under the Python License. It was commercialized initially by DeLano Scientific LLC, which was a private software company dedicated to creating useful tools that become universally accessible to scientific and educational communities. It is currently commercialized by Schrödinger, Inc. PyMOL can produce high-quality 3D images of small molecules and biological macromolecules, such as proteins. According to the original author, by 2009, almost a quarter of all published images of 3D protein structures in the scientific literature were made using PyMOL.[citation needed]
PyMOL is one of a few open-source model visualization tools available for use in structural biology. The Py part of the software's name refers to that it extends, and is extensible by, the programming language Python.
PyMOL uses OpenGL Extension Wrangler Library (GLEW) and FreeGLUT, and can solve Poisson–Boltzmann equations using the Adaptive Poisson Boltzmann Solver.[2] PyMOL uses Tk for the GUI widgets; Native Aqua binaries are available for macOS through Schrödinger.
"PyMOL Molecular Graphics System". SourceForge.
"poissonboltzmann".
-----------
PyMOL is an OpenGL molecular graphics system written in Python.
PyMOL is a molecular graphics system with an embedded Python interpreter designed for real-time visualization and rapid generation of high-quality molecular graphics images and animations.
It is fully extensible and available free to everyone via the "Python"
license.
Although a newcomer to the field, PyMOL can already be used to generate
stunning images and animations with unprecedented ease. It can also
perform many other valuable tasks (such as editing PDB files) to assist
you in your research.
Features include:

------------
Installing:
www
visible.
|
|
| Original author(s) | Warren Lyford DeLano |
|---|---|
| Developer(s) | Schrödinger, Inc. |
| Initial release | 2000 |
| Stable release |
1.8.6.2 / 26 June 2017
|
| Development status | Active |
| Written in | C, C++, Python |
| Operating system | Cross-platform: macOS, Unix, Linux, Windows |
| Platform | IA-32, x86-64 |
| Available in | English |
| Type | Molecular modelling |
| License | Python[1] |
| Website | www |
PyMOL is computer software, a molecular visualization system created by Warren Lyford DeLano. It is user-sponsored, open-source software, released under the Python License. It was commercialized initially by DeLano Scientific LLC, which was a private software company dedicated to creating useful tools that become universally accessible to scientific and educational communities. It is currently commercialized by Schrödinger, Inc. PyMOL can produce high-quality 3D images of small molecules and biological macromolecules, such as proteins. According to the original author, by 2009, almost a quarter of all published images of 3D protein structures in the scientific literature were made using PyMOL.[citation needed]
PyMOL is one of a few open-source model visualization tools available for use in structural biology. The Py part of the software's name refers to that it extends, and is extensible by, the programming language Python.
PyMOL uses OpenGL Extension Wrangler Library (GLEW) and FreeGLUT, and can solve Poisson–Boltzmann equations using the Adaptive Poisson Boltzmann Solver.[2] PyMOL uses Tk for the GUI widgets; Native Aqua binaries are available for macOS through Schrödinger.
Contents
Nonfree binaries
On 1 August 2006, DeLano Scientific adopted a controlled-access download system for precompiled PyMOL builds (including betas) distributed by the company. Access to these executables is now limited to registered users who are paying customers; educational builds are available free to students and teachers. However, most of the current source code continues to be available for free, as are older precompiled builds. While the build systems for other platforms are open, the Windows API (WinAPI, Win32) build system is not, although unofficial Windows binaries are available online.[3] Anyone can either compile an executable from the source code or pay for a subscription to support services to obtain access to precompiled executables.Acquisition by Schrödinger
On 8 January 2010, Schrödinger, Inc. reached an agreement to acquire PyMOL. The firm assumed development, maintenance, support, and sales of PyMOL, including all then-valid subscriptions. They also continue to actively support the PyMOL open-source community.Gallery
-
Example of some molecule editing features of PyMOL, dihedral bond rotation and interactive molecular relaxation with Sculpting mode. These are useful features to prepare input geometry for quantum chemistry software
-
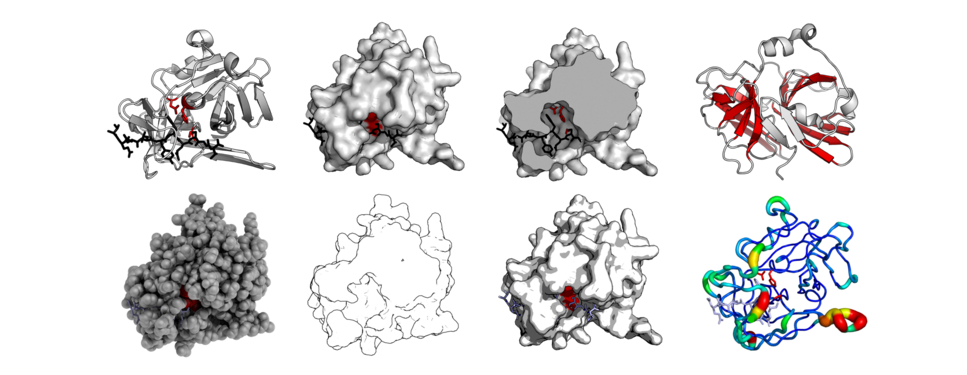
The same protein structure (TEV protease - PDB: 1LVB) rendered in different modes. Standard cartoon, surface, cut-through of surface, highlighted barrels, 'QuteMol'-like, 'Goodsell'-like, glossy-surface, and b-factor putty.
See also
- Abalone
- Comparison of software for molecular mechanics modeling
- Gabedit
- List of molecular graphics systems
- Molden
- Molecular modelling
- Molekel
- RasMol
- SAMSON
References
External links
| Wikimedia Commons has media related to PyMOL. |
-----------
PyMOL is an OpenGL molecular graphics system written in Python.
PyMOL is a molecular graphics system with an embedded Python interpreter designed for real-time visualization and rapid generation of high-quality molecular graphics images and animations.
| PyMOL 1.7.6.0 |
|
Price Free to download Size 8.3MB License Open-source, user-sponsored Developer DeLano Scientific LLC Website www.pymol.org
System Requirements
Support:3D OpenGL graphics acceleration Python tcl tk libpng zlib glut glut-devel pmw numeric Wiki, Tutorials, eMovie, PyMOL Plugins, Cams PyMOL Page
Selected
Reviews:
wordpress |
- Real-Time 3D visualization
- Publication quality renderings
- Extensive animation capabilities
- Support for X-ray crystallography
- Modular Architechture
- Volume visualization for unique display of volumetric data and simultaneous visualization of multiple iso-surfaces
- Electron density map generation from reflection data
- Stereochemical labeling
- Atom Typing; MacroModel and SYBYL/MOL2 support
- OpenGL Shaders (GLSL) for improved on-screen rendering (cartoons, sticks, surfaces, volumes)
- Dynamic measurements (distances, bond angles and dihedral angles)
- Flexible API for custom applications
- Support for bg_gradient when ray tracing
- Many non-standard nucleic acids (PSU, 5MU, 7MG, MIA, H2U, 4SU, MA6, UR3, 5MC, 40C, 2MG, TM2, 2MG) render correctly as cartoons
- Surface color smoothing
- Frame buffer-based antialiasing for real-time rendering
- Optimized anaglyph colors and intensities for an improved 3D viewing experience
- Real-time and ray-traced ambient occlusion
- Support for reading compressed Maestro (.maegz)** and PyMOL session files (.pze, pzw, .pse.gz, .psw.gz)
- Written in C and Python

------------
Installing:

No comments:
Post a Comment